About Me
Welcome to my personal website!

Life Science Researcher
An enthusiastic scientist with a strong passion about viral evolution and the application of vaccine design. Given the importance of bioinformatics in biomedical science research, I began my transition from bench-work (molecular biology/structural biology) to bioinformatics in 2017. I am interested in better understanding how vaccines/drugs work towards a more effective prevention/clearance of viral diseases, and how to overcome diseases with no or lack of an effective prophylactic or therapeutic intervention. Currently, I embark on my Ph.D. journey in Germany under COVIPA consortium that focus on gathering the knowledge about virological and immunological drivers of COVID-19 pathogenesis.
- Degree: Master
- Email: lichuinchong@gmail.com
- Interest: Virology, Vaccineinformatics, Immunoinformatics
- ResearchGate: Click HERE
- Google Scholar: Click HERE
- GitHub: Click HERE
Resume
Here is a short description of my journey.
Education
Doctor of Philosophy (Ph.D.)
Apr 2022 - Present
Hannover Biomedical Research School, Hannover Medical School, DE
Dissertation: Increasing pandemic preparedness by computational high-throughput virus discovery: Identification of RNA viruses and host reservoirs with high spillover risk
Supervisor: Prof. Dr. Chris Lauber
Master of Science (M.Sc.) Bioinformatics [by Research]
Sep 2018 - Sep 2020
School of Data Sciences, Perdana University, MY
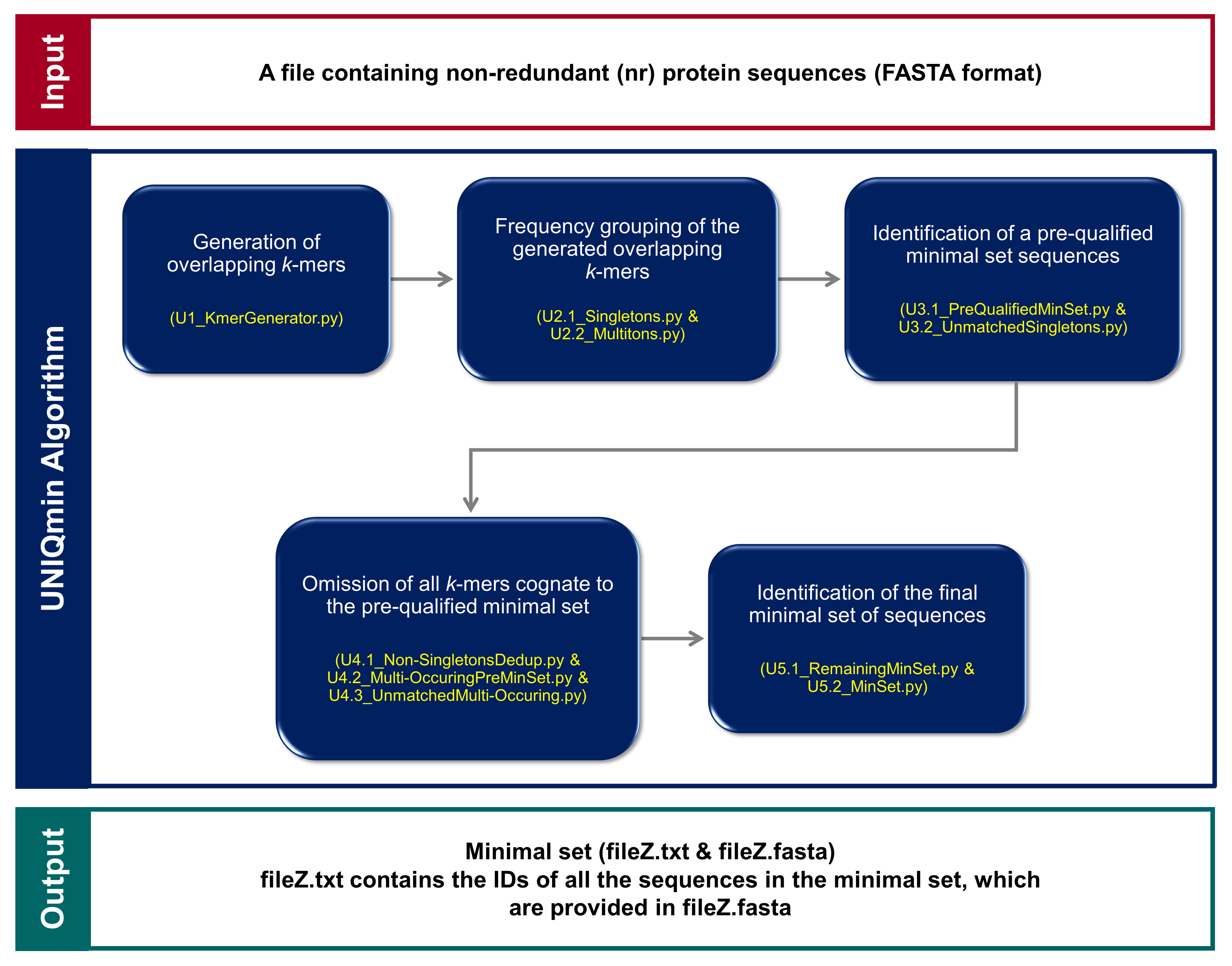
Dissertation: Mapping the minimal set of the viral peptidome
Supervisor: Assoc. Prof. Dr. Mohammad Asif Khan
Project Overview:
This project is aimed to elucidate the important insights of viral diversity from the minimal set of viral protein sequences by developing a more vigorous algorithm that would be
scalable to the massive dataset of all reported viruses in nature (~4.9M sequences as of Nov 2018).
Bachelor of Science (B.Sc.) Biomedical Sciences
Sep 2014 - Aug 2018
Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, MY
Honours Dissertation: Expression, purification and characterisation of the protuding
domain of Macrobrachium rosenbergii nodavirus (MrNV) capsid protein
Supervisor: Assoc. Prof. Dr. Ho Kok Lian
Project Overview:
This project, involving molecular and structural virology, is focused on producing and characterising the P-domain of MrNV
in order to provide further undertsanding of virus-host interactions and the viral pathogenicity.
Professional Experience (Selected)
Doctoral Researcher
Apr 2022 - Present
TWINCORE Centre for Experimental and Clinical Infection Research GmbH, DE
under the COVIPA consortium
Visiting Trainee
Oct - Nov 2023
Division of Virus Associated Carcinogenesis (F170), German Cancer Research Center (DKFZ), DE
under the Helmholtz Visiting Researcher Grant from Helmholtz Information & Data Science Academy (HIDA) Trainee Network
International Visiting Researcher
Mar - Sep 2021
Beykoz Institute of Life Sciences and Biotechnology (BILSAB), Bezmialem Vakif University, TR
under the 2232 International Fellowship for Outstanding Researchers Program of TÜBİTAK (Project No: 118C314)
Research Assistant
Aug - Dec 2020
Faculty of Dentistry, University of Malaya, MY
under Tabung Kursi Dr. Siti Hasmah Mohd Ali
Dec 2019 - May 2020
School of Data Sciences, Perdana University, MY
under Malaysian Medical Association (MMA) Research Grant
Sep 2018 - Mar 2019
School of Data Sciences, Perdana University, MY
under Adnuri SMA Research Grant
Research Projects
With the advance of high-throughput technology, omics data are playing an prominent role in the new phase of preventive medicine research.
As such, the integration of biomedical science knowledge with bioinformatics/data science skill is the general approach of my intended research direction.
Keywords: Virus, Virology, Proteomics, Bioinformatics, Vaccineinformatics, Immunoinformatics
- Ongoing Project
- All
- Vaccineinformatics
- Machine Learning
- Virology
In silico analysis of genetic variants in biological pathways associated with risk of gout
Collaboration with Dr. Tan Lay Kim from National Institutes of Health (NIH), Malaysia
pathway analysis
gene ontology
gout
Use the published gout-associated single nucleotide polymorphisms (SNPS) to identify the candidate genes and pathways that involved in the pathogenesis of gout.
Exploring the chemical space of SARS-CoV-2 drugs via machine learning
Under supervision of Assoc. Prof. Dr. Chanin Nantasenamat (Mahidol University, TH)
SARS-CoV-2
chemical space analysis
machine learning
Explore the chemical diversity of compounds against the various COVID-19 target proteins, including main protease, 3CL protease, spike protein etc.
Mapping HLA-supertype-restricted T-cell epitopes in SARS-CoV-2 proteome
Under supervision of Assoc. Prof. Dr. Mohammad Asif Khan (Perdana University, MY / BVU, TR)
SARS-CoV-2
T-cell epitope
HLA supertype
epitope-based vaccine candidate
sequence diversity
Performe a large-scale, proteome-wide mapping and diversity analyses of putative HLA supertype-restricted T-cell epitopes of SARS-CoV-2.
Mapping the minimal set of the viral peptidome
Under supervision of Assoc. Prof. Dr. Mohammad Asif Khan (Perdana University, MY / BVU, TR)
alignment-free
minimal set
UNIQmin
virus
viral diversity
Automated an alignment-independent algorithm as a tool, UNIQmin, to generate a minimal set of sequences from a given dataset, in order to elucidate the important insights of viral sequence diversity.
Identification of serotype-specific, highly conserved dengue virus sequences: Implications for vaccine design
Under supervision of Assoc. Prof. Dr. Mohammad Asif Khan (Perdana University, MY / BVU, TR)
dengue virus
serotype-specific
immune targets
vaccine design
mutual information
Performed a large-scale systematic study to map and characterise HCSS sequences in the DENV proteome, as candidates for vaccine target selection.
Expression, purification and characterisation of the protruding domain of Macrobrachium rosenbergii nodavirus capsid protein
Under supervision of Assoc. Prof. Dr. Ho Kok Lian (University Putra Malaysia, MY)
viral structure
recombinant protein
protein sequencing
protein expression
Produced and characterised the protuding domain of MrNV capsid protein in order to provide fruther undertsanding of virus-host interactions and the viral pathogenicity.
Avian influenza H7N9 virus adaptation to human hosts
Under supervision of Assoc. Prof. Dr. Mohammad Asif Khan (Perdana University, MY / BVU, TR)
influenza virus
zoonosis
avian viruses
human viruses
host adaptation
viral diversity
Performed a large-scale, quantitative analyses on all the changes or substitutions in the reported avian and human H7N9 protein sequencess.
Research Tools
These are the tools that I have developed or contributed throughout my journey of continuous learning.
- Ongoing Project
- All
- Bioinformatics
- Proteomics
- Vaccineinformatics
- Virology
Publications
as of April 2024
Citations
Papers Published
2024
Consensus Statement from the First RdRp Summit: Advancing RNA Virus Discovery at Scale across Communities
J Charon, I Olendraite, M Forgia LC Chong, et al
Frontiers in Virology
Perspective Article
2023
Machine Learning Approaches to Study the Structure-activity Relationships of LpxC Inhibitors
T Yu, LC Chong, C Nantasenamat, N Anuwongcharoen, T Piacham*
EXCLI Journal
Original Research
Viroid-like RNA-dependent RNA Polymerase-encoding Ambiviruses are Abundant in Complex Fungi
LC Chong, C Lauber*
Frontiers in Microbiology
Original Research
Genomic Analysis and Biological Characterization of A Novel Schitoviridae Phage Infecting Vibrio alginolyticus
S Tajuddin, AM Khan*, LC Chong, et al
Applied Genetics and Molecular Biotechnology
Original Research
2022
Historical Milestone in 42 Years of Viral Sequencing — Impetus for A Community-driven Sequencing of Global Priority Pathogens
LC Chong, AM Khan*
Frontiers in Microbiology
Opinion Article
Negligible Peptidome Diversity of SARS-CoV-2 and Its Higher Taxonomic Ranks
LC Chong, AM Khan*
bioRxiv
Original Research
UNIQmin, An Alignment-free Tool to Study Viral Sequence Diversity across Taxonomic Lineages: A Case Study of Monkeypox Virus
LC Chong, AM Khan*
bioRxiv
Protocol Article
The Third International Hackathon for Applying Insights into Large-scale Genomic Composition to Use Cases in A Wide Range of Organisms
K Walker*, D Kalra*, R Lowdon*, …, LC Chong, et al
F1000Research
Software Tool Article
iCn3D: From Web-Based 3D Viewer to Structural Analysis Tool in Batch Mode
J Wang*, P Youkharibache, A Marchler-Bauer, …, LC Chong, et al
Frontiers in Molecular Biosciences
Original Research
Role of Probiotics in the Management of COVID-19: A Computational Perspective
QV Nguyen, LC Chong, YY Hor, LC Lew, IA Rather* & SB Choi*
Nutrients
Review
2021
An Alignment-independent Approach for the Study of Viral Sequence Diversity at Any Given Rank of Taxonomy Lineage
LC Chong, WL Lim, KHK Ban, AM Khan*
Biology
Original Research
Drug Discovery of Spinal Muscular Atrophy (SMA) from the Computational Perspective: A Comprehensive Review
LC Chong, G Gandhi, JM Lee, WWY Yeo, SB Choi*
International Journal of Molecular Sciences
Review
Perceived Stress of Quarantine and Isolation During COVID-19 Pandemic: A Global Survey
TGC Collaborative
Frontiers in Psychiatry
Original Research
Avian Influenza H7N9 Virus Adaptation to Human Hosts
S Tan, MF Sjaugi, SC Fong, LC Chong, HS Abd Raman, NEN Mohamed, JT August, AM Khan*
Viruses
Original Research
Psychological Impacts and Post-traumatic Stress Disorder among People under COVID-19 Quarantine and Isolation: A Global Survey
TG COVID
International Journal of Environmental Research and Public Health
Original Research
2019
Identification of Highly Conserved, Serotype-specific Dengue Virus Sequences: Implications for Vaccine Design
LC Chong, AM Khan*
BMC Genomics
Original Research
Expression, Purification and Characterization of the Dimeric Protruding Domain of Macrobrachium rosenbergii Nodavirus Capsid Protein Expressed in Escherichia coli
LC Chong, H Ganesan, CY Yong, WS Tan, KL Ho*
PloS one
Original Research
Vaccine Target Discovery
LC Chong, AM Khan*
Encyclopedia of Bioinformatics and Computational Biology
Book Chapter